Managing domains#
The geometrical definition of a domain consists in a set of geometrical elements (such as triangles, hexahedra, prisms, …) that are related to a mesh
handled through a Mesh object containing the geometrical domains through GeomDomain objects.
Generally, users handle Domain alias of GeomDomain (more precisely a reference to).
Geometrical domains are fundamental objects because they are the support of integrals or boundary conditions involved in variational problem. These domains are defined by mesh tools, using names and sidenames in definition of geometries or given as physical domain in .geo file.
Retrieving domains#
In order to be used in program, the domains have to be ’retrieved’ as Domain object from mesh:
Strings sn("y=0", "x=1", "y=1", "x=0");
Mesh mesh2d(SquareGeo(_origin=Point(0.,0.), _length=1, _nnodes=20, _side_names=sn), _generator=structured);
Domain omega=mesh2d.domain("Omega");
Domain sigmaM=mesh2d.domain("x=0");
Domain sigmaP=mesh2d.domain("x=1");
Domain gammaM=mesh2d.domain("y=0");
Domain gammaP=mesh2d.domain("y=1");
By default, “Omega” is the string name of the main domain of mesh. A domain of a mesh can be renamed:
Strings sn("", "x=1/2−", "", "x=0");
Mesh mesh2d(Rectangle(_origin=Point(0.,0.), _xlength=0.5, _ylength=1, _nnodes=Numbers(20,40), _side_names=sn), _generator=structured);
mesh2d.renameDomain("Omega","Omega−");
sn[1] = "x=1"; sn[3] = "x=1/2+";
Mesh mesh2d2(Rectangle(_origin=Point(0.5,0.), _xlength=0.5, _ylength=1, _nnodes=Numbers(20,40), _side_names=sn), _generator=structured);
mesh2d2.renameDomain("Omega","Omega+");
mesh2d.merge(mesh2d2);
Domain omegaM=mesh2d.domain("Omega−");
Domain omegaP=mesh2d.domain("Omega+");
Domain sigmaM=mesh2d.domain("x=0");
Domain sigmaP=mesh2d.domain("x=1");
Domain gamma=mesh2d.domain("x=1/2−_x=1/2+");
In this example, the unit square is split in two domains \(\Omega^+\) and \(\Omega^-\) using the merge and renameDomain functions.
Note that the merging process of meshes concatenates ’same’ domain (“x=1/2-” and “x=1/2+”) in a new one named “x=1/2-_x=1/2+”.
Warning
A Domain is a reference to a GeomDomain object. As a consequence, you can not instanciate, for instance, a std::vector<Domain>.
This is the reason why there exist the Domains class to deal with a vector of Domain.
Global operations on domains#
There exists two set operations: the union of domains (merge function and + operator) and the difference of two domains (- operator).
Union of domains#
The merge function accepts up to 6 domains. When you want to merge only 2 domains, you may use the + operator, which is more readable.
Rectangle rect(_origin=Point(0.,0.), _xlength=1., _ylength=1., _nnodes=31, _domain_name="Omega", _side_names={"Gamma1","Gamma2","Gamma3","Gamma4"});
Mesh mrect(rect);
Domain omega=mrect.domain("Omega"),
gamma1=mrect.domain("Gamma1"),
gamma2=mrect.domain("Gamma2"),
gamma3=mrect.domain("Gamma3"),
gamma4=mrect.domain("Gamma4");
Domain gamma12 = merge(gamma1, gamma2); // union of elements of gamma1 and gamma2
Domain gamma34 = gamma3 + gamma4; // union of elements of gamma1 and gamma4
Domain gamma = merge(gamma1, gamma2, gamma3, gamma4, "gamma"); // union of all side domains
Hint
When more than 2 domains are involved in a merge operation, it is more efficient to use the merge function than to use the + operator, producing intermediate domains.
Domain of sides of elements#
It may be useful to get the domain of all sides (points, edges, faces) of an existing domain. For instance the set of all faces of a volumic domain, the set of all edges of a surfacic domain. The sides function or the GeomDomain::sidesDomain member function do the job:
SquareGeo sq(_origin=Point(0.,0.), _length=1., _nnodes=2, _domain_name="Omega", _side_names="Gamma")
Mesh msq(sq, _generator=structured);
Domain omega=msq.domain("Omega"), gamma=msq.domain("Gamma");
Domain somega=omega.sidesDomain(); // edges of square domain
Domain somega2 = sides(omega); // alternate call
Domain sgamma=gamma.sidesDomain(); // edges of square boundary domain
Domain sgamma2=sides(gamma); // alternate call
This works for every geometry dimension. Here is a 3D example:
Mesh mcu(Cube(_origin=Point(0.,0.,0.), _length=1., _nnodes=2, _domain_name="Omega"), _shape=hexahedron, _generator=structured);
Domain omega=mcu.domain("Omega");
Domain somega=sides(omega); // faces of cube domain
See also
Applying GeomDomain::sidesDomain on a side domain, you get the sides of sides:
Mesh mcu(Cube(_origin=Point(0.,0.,0.), _length=1., _nnodes=2, _domain_name="Omega"), _shape=hexahedron, _generator=structured);
Domain omega=mcu.domain("Omega");
Domain somega=sides(omega); // faces of cube domain
Domain ssomega=sides(somega); // edges of cube domain
By default, the name of domain is “sides of domain.name()”, by specifying a string as argument of GeomDomain::sidesDomain you can change it.
Difference of domains#
The difference of two domains is defined as the set of elements of the first domain that are not in the second one. It is done using the - operator:
Rectangle rect(_origin=Point(0.,0.), _xlength=1., _ylength=1., _nnodes=31, _domain_name="Omega", _side_names={"Gamma1","Gamma2","Gamma3","Gamma4"});
Mesh mrect(rect);
Domain omega=mrect.domain("Omega"),
gamma1=mrect.domain("Gamma1"),
gamma2=mrect.domain("Gamma2"),
gamma3=mrect.domain("Gamma3"),
gamma4=mrect.domain("Gamma4");
Domain gamma = merge(gamma1, gamma2, gamma3, gamma4, "gamma"); // union of all side domains
Domain sigma = sides(omega); // set of all sides of elements of omega
Domain sigma0 = sigma - gamma; // set of all internal sides of elements of omega
See also
By using the internalSides function or the GeomDomain::internalSidesDomain member function, it is also possible to get the domain of internal sides, i.e. the union of all sides excluding the sides located on the boundary (in other words the union of shared sides).
Dealing with normals#
Normal vectors may be required in many variationnal forms, in particular when using BEM like methods. In (bi)linear forms, they appear with symbolic names _n, _nx, _ny that corresponds to real normal vectors. The question is which normal vectors are selected by XLiFE++.
Note that the normal vectors of a domain, say \(\Gamma\), are computed only if the domain is a manifold, say a surface/curve domain in a 3d/2d space. If they are required, they are automatically computed with the following default rules:
If \(\Sigma\) is a boundary (or a part of) of a unique domain \(\Omega\), the selected normals are the outwards vectors to \(\Omega\);
if \(\Sigma\) is a boundary between two domains (an interface), the selected normals are the towards infinite vectors;
if \(\Sigma\) is not a boundary, say an immersed manifold, the selected normals are the towards infinite vectors.
The user can modify the normal orientation by using the member function of Domain class: setNormalOrientation(OrientationType[, Domain]) where OrientationType has one of the following values (default is _undefOrientationType):
_towardsInfinite, _outwardsInfinite // for any side domain
_towardsDomain, _outwardsDomain // for any boundary or interface
To change the normal orientation of a side domain Sigma, write for instance
Sigma.setNormalOrientation(_towardsInfinite); // towards infinite normals
Sigma.setNormalOrientation(_outwardsDomain); // outwards normals
// UNSAFE for an interface!
Sigma.setNormalOrientation(_outwardsDomain, Omega);//outwards normals to Omega
Attention
All normal vectors of an open manifold are oriented consistently, but the global orientation is unpredictable because it depends on the first element encountered. Check the orientation by visualizing the normal vectors (see below).
The global orientation may be controlled by using a virtual “interior” point that orients the normal vector related to this interior point:
Sigma.setNormalOrientation(_towardsInfinite, Point(0,0));
This trick allows dealing with simple open manifolds, but for complex geometries (union of open manifolds for instance) the global orientation of normal vector remains uncontrollable.
For visualization purpose, the normal vectors can be exported to a vtk file using:
Sigma.saveNormalsToFile("n_Sigma", vtk);
They can be also collected in a TermVector object:
Space V1(_domain=Omega, _interpolation=P1, _name="V1");
Unknown u1(V1, _name="u1", _dim=3);
TermVector Ns = normalsOn(Sigma, u1);
The normals are computed using L2 projection on the space \(W\) related to the Unknown used. More precisely, the normals \(n\) in space \(W\) solves the problem:
where \(n_0\) is the normal to the element faces. If \((wj)\) denotes the basis of \(W\) and \(N\) the vector representing the normal in \(W\) basis ( \(n=\Sigma n_j\cdot w_j\)), the following linear system is solved:
The unknown may be an explicit vector unknown or a scalar unknown if the space is a space of vectors. In the case of a P0 unknown, the normals are normals on element faces with no projection. In the case of Pk Lagrange unknown, the normals are some interpolated normals on Lagrange dofs.
Map of domains#
Some processes require a geometric map between two domains. For instance, to deal with periodic condition related to two side domains:
The elimination process uses the geometric map \(F:\ \ \Sigma^+ \longrightarrow \Sigma^-\). The simple way to define such map is the following:
Vector<Real> mapPM(const Point& P, Parameters& pa = defaultParameters)
{
Point Q(P);
Q(1)-=1;
return Q;
}
...
defineMap(sigmaP, sigmaM, mapPM);
Caution
The mapPm function returns a Vector<Real> (alias Reals) which is more general than a Point. Please respect this prototype!
Hint
When dealing with curved domains, generally meshed with P1,P2, … elements, you can use some natural maps from one domain to the other (think about the scaling transformation of a circle to another circle whereas the circles are approximated by polygons). In that case, points are transported using the given geometrical transformation and an additional projection to guarantee that the image points belong to the arrival domain. Obviously, the projection induces an error related to the mesh size.
Warning
It is currently not possible to define two different maps for a pair of domains.
Assign properties to domains#
In some problems, physical properties may be different from a domain to other one. This may be managed by differentiating integrals in variational formulation:
But it may be too intricate if there are a lot of domains or integrals. So there is an alternative method consisting in defining a unique function \(rho\) and deal with a unique integral:
and assign id to domains that are subdomains:
Real rho(const Point&P, Parameters& pars=defaultParameters)
{
Number mat=getMaterialId();
if(mat==1) return 1.;
return 2.;
}
...
Domain omega=mesh2d.domain("omega"); // whole domain
Domain omega1=mesh2d.domain("omega_1"); // subdomain
Domain omega2=mesh2d.domain("omega_2"); // subdomain
omega1.setMaterialId(1);
omega2.setMaterialId(2);
Dealing with crack#
There exist two ways in XLiFE++ to insert cracks. The first one consists in using the Gmsh capabilities and the other one consists in using the intern cracking tool that it is attached to the Mesh class.
Cracking using Gmsh capabilities#
Gmsh allows to crack domains (1D cracks in 2D meshes, 1D or 2D cracks in 3D meshes). Cracks can be opened (boundary nodes of the crack are duplicated as the other nodes) or closed crack (boundary nodes of the crack are not duplicated).
There is a general routine defining both opened and closed cracks through 2 additional optional arguments: Geometry::crack, default behavior being closed cracks.
As an alternative, it can be used the functions closedCrack (only the geometry in argument) and openCrack (the geometry and a domain name) to define a closed or an opened crack.
The domain name is the boundary domain of the geometry to be cracked (opened). For instance :
Point x1(0,0,0), x2(1,0,0), x3(1,1,0), x4(0,1,0), x5(0.2,0.2,0), x6(0.8,0.8,0)
Rectangle rrect8(_v1=x1, _v2=x2, _v4=x4, _domain_name="Omega", _side_names="Gamma");
Segment scrack(_v1=x5, _v2=x6, _nnodes=3, _domain_name="Crack", _side_names="Sigma");
openCrack(scrack, "Sigma");
Mesh m(rrect8+scrack, _generator=gmsh);
A side name is given to both ends of the segment. This name will be given to the routine openCrack to tell which ends are to be opened.
Fig. 53 Open crack.
Fig. 54 Open crack.
See Cracking geometries for more details.
Cracking using internal tool#
The way to define a crack is very similar to the previous one, except the crack function acts on a mesh and a side domain (will be the crack) and not on the geometry:
Point x1(0,0,0), x2(1,0,0), x3(1,1,0), x4(0,1,0), x5(0.2,0.2,0), x6(0.8,0.8,0), x7(0.2,0,0), x8(0.8,1,0);
Rectangle r(_v1=x1, _v2=x2, _v4=x4, _domain_name="Omega", _side_names="Gamma");
Segment s(_v1=x5, _v2=x6, _nnodes=3, _domain_name="Sigma");
Mesh m(r+s, _generator=gmsh); //mesh as usual
Domain sigma=m.domain("Sigma"); //get the crack domain
m.crack(sigma); //mesh is now cracked
By default, the crack is not open (end points are not duplicated). By specifying _opencrack when calling the crack function, the crack will be open (end points are duplicated):
m.crack(sigma, _openCrack);
Once the mesh is cracked, the sides of the crack are named xxx+ and xxx- where xxx is the name of the original domain supporting the crack. It is possible to crack a domain with few segments, naming the segments with the same name. Even, one can crack a closed domain (for instance a polygon).
Warning
This tool is working in 2D and 3D, but fails when trying to crack a domain with a triple point (for instance an Y shape crack).
Mesh quality statistics#
To known statistics about the mesh, the minimum, maximum and average value of :
measure and size of elements
measure of side of elements
call computeStatistics on domains of interest:
Domain omega=...
omega.computeStatistics();
theCout << omega << eol;
computeStatistics have an optional boolean argument, whose default value is false, meaning deactivation of the computation of side statistics.
Partitioning a domain#
XLiFE++ can partition a mesh by partitioning its geometric domain (Omega) with the function partition using Metis.
This partitioning can be built by the function partition from a Domain object:
Mesh m(...);
Domain omega = m.domain("Omega");
omega.partition(_nbParts=4, _partmesh="Dual", _ncommon=2, _iptype=1, _ptype=0,
_objtype=0, _ctype=0, _rtype=1);
Hint
The essential key is _nbParts, setting the number of partitions, but most options are proposed with default value (bold in the tab):
|
associated graph partitioning method |
|
|
number of common nodes between 2 elements |
|
|
algorithm for initial partitioning |
|
|
Partitionning method |
|
|
goal |
|
|
schema for mesh coarsening |
|
|
algorithm for refinement |
|
Warning
All options are not compatible with each other for obvious reasons. For example:
_ptype=recursive with _objtype=Vol or _iptype=random with _ptype=Kway.
Furthermore, information about partition can be obtained with the access fonction partitionData.
For example,
|
number of interface sides. |
|
balance. |
|
Objval. |
At last, elements and vertices of each part are stored in maps:
std::map<Number, Int> elementsByPartition_mstd::map<Number, Int> verticesByPartition_m
and, elements and vertices of each interface are stored in lists:
std::list<Number> interfaceVertices_lstd::list<Number> interfaceElements_l
Example
Partition of a mesh in 10 parts with options:
NbParts=10 |
Ncommon=2 |
and default values:
partMesh=Dual |
Rtype=FM |
Ctype=RM |
Ptype=Recursive |
Objtype=Cut |
IPtype=Grow |
Mesh m(...);
Domain omega = m.domain("Omega");
omega.partition(_nbParts=10, _ncommon=2);
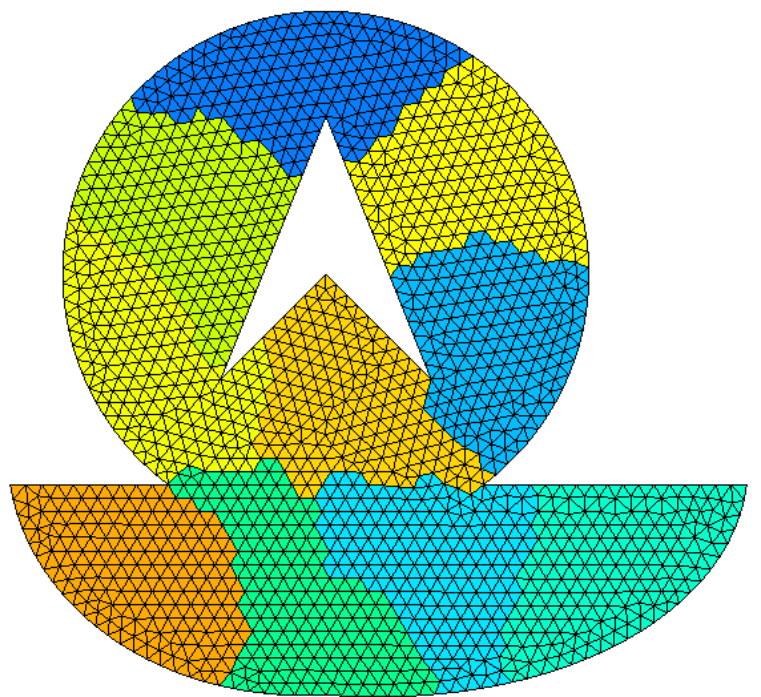
Summary of domain operations#
We only list here the stuff interesting users:
Properties |
|
|---|---|
|
number of elements in domain |
|
set normal orientation of a side domain |
|
access to parametrization if defined |
|
associates a material id |
|
associate a domain id |
|
measure of a domain |
|
relates |
Domain building |
|
|
domain of sides of elements of |
|
domain of internal sides of elements of |
|
crack domain from both sides of the crack |
|
extension domain of a side domain |
|
fictitious domain of a side domain |
|
union of n domains with same element dimension |
|
union of 2 domains with same element dimension |
|
difference of 2 domains |
Dealing with parametrizations#
XLiFE++ may associate a Parametrization object to each Geometry object. It is accessible using the member function parametrization:
Disk di(_center=Point(0.,0.,0.),_radius=1.,_nnodes=20);
const Parametrization& par=di.parametrization();
const Parametrization& par=di.boundaryParametrization();
Parametrization exists for any canonical geometry and also for composite geometry, but in that case it is a PiecewiseParametrization object (inheriting from Parametrization).
It is also possible to access to the parametrization of the boundary.
Parametrization class offers access to some useful geometrical properties (derivatives, jacobian, curvilinear abscissa curvatures, normal and tangent vectors, …); see Parametrization documentation.
Parametrization objects are attached to the geometry of a GeomDomain object, but they can be accessed straightforward using the same functions:
Disk di(_center=Point(0.,0.,0.), _radius=1., _nnodes=20, _domain_name="Omega");
Mesh m(di, _generator=gmsh);
Domain omega=m.domain("Omega");
const Parametrization& par=omega.parametrization();
const Parametrization& par=omega.boundaryParametrization();
Warning
Parametrizations are not yet available for geometries coming from OpenCascade.
This preliminary part is detailed in chapter Mesh generation.
